3. Connections
Formulation
- Mapping the network of connection
Model Definition
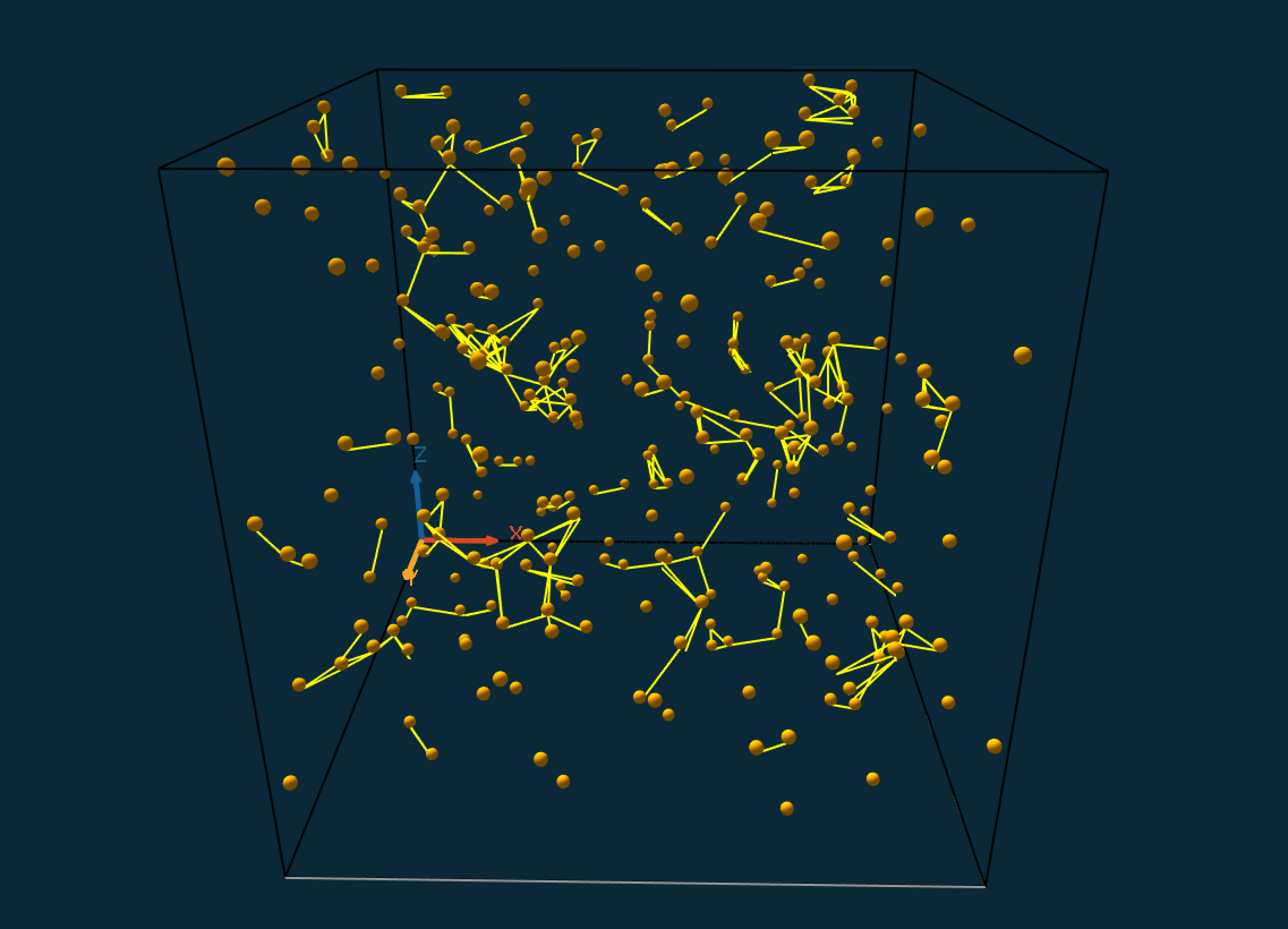
In this final step, we will display edges between cells that are within a given distance.
Cells update
We add a new reflex to collect the neighbors of the cell that are within a certain distance:
species cells skills:[moving3D]{
...
reflex compute_neighbors {
neighbors <- cells select ((each distance_to self) < 10);
}
}
Then we update the cell aspect as follows. For each element (cells) of the neighbors list, we draw a line between this neighbor's location and the current cell's location.
aspect default {
draw sphere(environment_size*0.01) color: #orange;
loop pp over: neighbors {
draw line([self.location,pp.location]);
}
}
Complete Model
The GIT version of the model can be found here Model 03.gaml
model Tuto3D
global {
int nb_cells <- 100;
int environment_size <- 100;
geometry shape <- cube(environment_size);
init {
create cell number: nb_cells {
location <- {rnd(environment_size), rnd(environment_size), rnd(environment_size)};
}
}
}
species cell skills: [moving3D] {
rgb color;
list<cell> neighbors;
int offset;
reflex move {
do wander;
}
reflex compute_neighbors {
neighbors <- cell select ((each distance_to self) < 10);
}
aspect default {
draw sphere(environment_size * 0.01) color: #orange;
loop pp over: neighbors {
draw line([self.location, pp.location]);
}
}
}
experiment Tuto3D type: gui {
parameter "Initial number of cells: " var: nb_cells min: 1 max: 1000 category: "Cells";
output {
display View1 type: opengl background: rgb(10, 40, 55) {
graphics "env" {
draw cube(environment_size) color: #black empty: true;
}
species cell;
}
}
}